Native
chromatin immunoprecipitation (N-ChIP) of Schistosoma mansoni (18/02/2014)
version 3
© Christoph Grunau,
Céline Cosseau, Abdelhalim Azzi, Jerome Buard, Julie Lepesant, David Roquis,
Sara Fneich, Marion Picard (2014)
This protocol is based on the protocol of David
Umlauf.
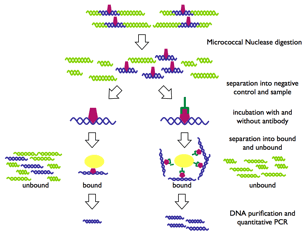
Figure 1: General scheme
Before
everything begins: prepare solutions...
· 1 M
KCl, autoclave
· 5 M
NaCl, filter and autoclave
· 1 M
MgCl, autoclave
· 1 M
Tris/Cl pH 7.4 - 7.6, autoclave
· 0.5
M EDTA, autoclave
· 1 M
CaCl2, 10 ml, sterile filter
· 100
mM DTT, 1ml (store at -20°C)
· Roche
CompleteProtease Inhibitor (ref: 11 697 498 001)
· 2.5
M Sodium butyrate (Sigma B5887 1g) (store at 4°C)
· 25
mM PMSF in isopropanol, 10 ml (store at -20°C)
· 15
U/µl Micrococccal nuclease (MNase) (USB 70196Y) in sterile 50% glycerol,
aliquot to ~10µl and store at -20°C
· Protein
A - sepharose CL-4B (Sigma P3391 250mg) (store at 4°C)
· agarose
gel loading buffer
· 20%
SDS, sterile filter
· 20
g/l glycogen solution (store at -20°C)
· 2%
NaN2 in water (store at 4°C), NaN2 is very toxic!
·
micro-dialysis units
(Slide-a-Lyzer 3500 D cut-off, Pierce 69550). Note: You can also make your own
(much cheaper) micro-dialysis units. For details see here
(external link, not tested).
|
preparation of protein A - sepharose |
|
1.
weight 250 mg protein A -
sepharose in 15 ml falcon tube 2.
wash with 10 ml sterile water 3.
centrifuge 10 min at 4000 rpm 4.
remove supernatant 5.
repeat step 3 - 4 four times 6.
add sterile water to 5 ml 250 mg Protein A - sepharose
swells to approx. 1 ml gel and binds approx. 20 mg human IgG. You will need
50 µl of the protein A sepharose homogeneously mixed in its 5 ml water volume
per ChIP. Only if prolonged storage is anticipated (several month) add NaN2
to 0.02 % and label tube appropriately. |
|
optional if micro dialysis units are not
available: preparation of dialysis tubing |
|
1.
cut tubing (e.g. VWR
international dialysis tube 0.5 mm) into pieces of 10 - 20 cm length 2.
boil for 10 min in a large
volume of 2% (w/v) sodium bicarbonate and 1 mM EDTA 3.
rinse the tubing thoroughly
with distilled water 4.
boil for 10 min in 1 mM EDTA 5.
cool and store in this
solution at 4°C 6.
before use, wash tubing
inside and outside with distilled water |
day 1...
· reserve
centrifuge and cool down to 4°C
· prepare
a 2% 0.5x TBE agarose gel with 20 µl slots
· preheat
a water bath to exactly 37°C
· prepare
the following solutions with autoclaved distilled water
|
2x base buffer |
||
|
6 ml |
1 M KCl |
60 mM final 1x |
|
0.3 ml |
5 M NaCl |
15 mM |
|
0.5 ml |
1 M MgCl2 |
5 mM |
|
20 µl |
500 mM EDTA |
0.1 mM |
|
1.5 ml |
1 M Tris/Cl |
15 mM |
|
to 50 ml |
water |
|
|
2 |
Roche protease inhibitor tabletts |
|
|
buffer1 (0.3 M sucrose) |
|
|
2.58 g |
sucrose |
|
12.5 ml |
2x base buffer |
|
50 µl |
sodium butyrate |
|
100 µl |
PMSF |
|
125 µl |
DTT |
|
to 25 ml |
water |
|
buffer 2 |
|
|
10 ml |
buffer 1 (0.3 M sucrose) |
|
put on 37°C to allow NP40 to be pipetted into the
buffer |
|
|
80 µl |
NP40 (cut pipette tip) |
|
put on 37°C to fully dissolve NP40 and put on ice |
|
|
buffer 3 (1.2 M sucrose) for 3 cell samples |
|
|
20.55 g |
sucrose |
|
25 ml |
2x base buffer |
|
100 µl |
sodium butyrate |
|
200 µl |
PMSF |
|
250 µl |
DTT |
|
to 50 ml |
water |
|
MNase digestion buffer |
|
|
1.1 g |
sucrose |
|
0.5 ml |
Tris/Cl |
|
80 µl |
PMSF |
|
40 µl |
MgCl2 |
|
20 µl |
sodium butyrate |
|
10 µl |
CaCl2 (essential for the enzyme) |
|
to 10 ml |
water |
|
put at 37°C |
|
|
Dialysis buffer |
|
|
1mM Tris/Cl, 200 µM EDTA, 200 µM PMSF, 5 mM sodium
butyrate |
|
|
50 µl |
Tris/Cl |
|
20 µl |
EDTA |
|
400 µl |
PMSF |
|
100 µl |
sodium butyrate |
|
to 50 ml |
water |
· put
all buffer solutions on ice (except MNase buffer)
· cell
lysis
o aliquote
1500 sporocysts, miracidia or 10-20 adults (stored at -80°C or in liquid
nitrogen)
o adults:
§ remove
excess liquid (if any) and resuspend in 1 ml buffer 1, add 1 ml buffer 2 (lysis
buffer) and transfer to Dounce
§ homogenize
for 3 min with Dounce (pestle A) on ice
§ put
on ice 7 min
o sporocysts
or miracidia:
§ centrifuge
into Eppendorf tubes, rinse storage tubes with PBS to recover all larvae
§ centrifuge
at 3400G, 10 min, 4°C
§ remove
supernatant
§ resuspend
completely in 1 ml buffer 1
§ do
not add human lymphoblast cells as carrier
§ add
1 ml buffer 2 (lysis buffer) and homogenize for 3 min with Dounce (pestle A) on
ice
§ put
on ice 7 min
o fill
8 ml buffer 3 into a 50 ml corex centrifugation tube
o overlay
the 8 ml buffer 3 with 1 ml cell suspension so that the tubes are ready for
centrifugation 15 min (sporocysts) of 10 min (adults) after buffer 2 has been
added to the cells
o disturb
a little bit the interface
o use
2 corex tubes for the sporocysts sample that is in 2 ml buffer 1+2
o mark
tubes at the exterior side (to know where to look for the nuclei)
o centrifuge
7800G 20 min 4°C
o carefully
remove supernatant completely (by pipetting with 10 ml pipette and then,
pipetting with micropipettes 1000µl>100µl>10µl)
· MNase
digestion
o resuspend
pellet in 1 ml MNase digestion buffer (2 ml for 10 000 miracidia or
cercariae)
o aliquot
500 µl of this suspension in 1.5 ml Eppendorf tubes
o add
1 µl MNase (15 U) and incubate 4 min at 37°C
o to
stop the reaction add 20 µl 0.5 M EDTA to each 500 µl MNase digest and put the
tube on ice
o centrifuge
13000 g 10 min 4°C
o transfer
the supernatant to a new tube (S1) and keep the pellet (P1)
o store
S1 at -20°C
o
Quantify chromatin in S1 with the Qubit® 2.0 Fluorometer (HS
DNA assay) following manufacturer instructions.
· Dialysis
of P1
o humidify
Slide-a-Lyzer with 50µl dialysis buffer
o resuspend
the pellet P1 in 100 µl dialysis buffer and dialyze overnight at 4°C against 50
ml dialysis buffer with gentle stirring
day 2...
o the
next day, transfer dialysed sample to Eppendorf tubes and...
§ centrifuge
13000 g 10 min 4°C
§ transfer
the supernatant to a new tube and repeat the centrifugation 2 times
§ supernatant
is fraction S2
o yesterdays
supernatant S1...
§ in parallel
with the dialyses sample, centrifuge 13000 g 10 min 4°C
§ transfer
the supernatant into a new tube and repeat this centrifugation 2 times
o these
triple centrifugations are IMPORTANT! They reduce the unspecific
background!
o use
50 µl of S1 and S2 for phenol/chlorofrom extraction, centrifuge and load 20 µl
of supernatant on 2% 0.5x TBE gel (100V, 25 min)
· incubation
with antibody
o Ideally,
the antibody should be in excess over the protein you want to precipitate. The
antigen/antibody ration must be determined experimentally for each antibody .
Prepare a dilution series of your chromatin in MNase buffer starting with 20 -
40 µg DNA for histon ChIP.
o Add
appropriate amounts of stock solutions to generate the
|
antibody incubation buffer |
|
|
NaCl |
150 mM |
|
Tris/Cl |
20 mM |
|
sodium butyrate |
20 mM |
|
EDTA |
5 mM |
|
PMSF |
100 µM |
o you
can download an Excel worksheet for calculation here (v0.1) or here (v1.0)
o dissolve
chromatin from S1 (and S2 if you have dialyzed) in 1 ml buffer
o add
about 2 µg antibody
o incubate
overnight at 4°C on a rotating wheel
day 3...
·
precipitation
o prepare
50 µl of protein A - sepharose for each tube
o wash
the beads with milliQ water in 1.5ml Eppendorf: centrifuge 4000G, 5 minutes,
remove supernatant and replace with equal volume of sterile water
o repeat
the washing two more times
o add
50 µl of protein A - sepharose to each tube
o incubate
at least 4 h at 4°C on a rotating wheel
o prepare
washing buffers (10 ml / tube) and cool down to 4°C:
|
washing buffers A B C |
50 ml |
100 ml |
200 ml |
300 ml |
||
|
|
Tris/Cl |
50 mM |
2.5 ml |
5 ml |
10 ml |
15 ml |
|
|
EDTA |
10 mM |
1 ml |
2 ml |
4 ml |
6 ml |
|
|
sodium butyrate |
5 mM |
100 µl |
200 µl |
400 µl |
600 µl |
|
washing buffer A |
NaCl |
75 mM |
750 µl |
1.5 ml |
3 ml |
4.5 ml |
|
washing buffer B |
NaCl |
125 mM |
1.25 ml |
2.5 ml |
5 ml |
7.5 ml |
|
washing buffer C |
NaCl |
175 mM |
1.75 ml |
3.5 ml |
7 ml |
10.5 ml |
o centrifuge
chromatin/antibody mixture 10 min 4°C 11600 g
o keep
the supernatant in a 2 ml tube. This is the unbound fraction UB.
o resuspend
the pellet in approx. 1 ml washing buffer A and transfer into a 15 ml Falcon
tube containing 9 ml washing buffer A
o mix
for 10 min on a rotating wheel at 4°C (speed 6)
o centrifuge
10 min 3400 g 4°C and pour off supernatant (by inversion)
o add
10 ml washing buffer B, mix for 10 min on a rotating wheel at 4°C and
centrifuge 10 min 4000 rpm 4°C
o pour
off supernatant (by inversion)
o add
10 ml washing buffer C, mix for 10 min on a rotating wheel at 4°C and
centrifuge 10 min 4000 rpm 4°C
o pour
off supernatant (by inversion)
o centrifuge
10 min 4000 rpm 4°C
o remove
remaining supernatant completely (not by inversion but by pipetting with
micropipettes 1000 µl, then 100 µl, then 10µl)
o resuspend
pellet in 500 µl elution buffer
|
elution buffer |
10 ml |
20 ml |
|
|
SDS (20% stock) |
1 % |
500 µl |
1ml |
|
Tris/Cl |
20 mM |
200 µl |
400 µl |
|
NaCl |
50 mM |
100 µl |
200 µl |
|
EDTA |
5 mM |
100 µl |
200 µl |
|
sodium butyrate |
20 mM |
80 µl |
160µl |
|
PMSF |
100 µM |
40 µl |
80 µl |
|
water |
|
to 10 ml |
to 20 ml |
o transfer
suspension to a 1.5 ml Eppendorf tube
o incubate
15 min at RT on a rotating wheel
o centrifuge
10 min 11600 g 18°C
o transfer
supernatant to a 1.5 ml Eppendorf tube
o This
is the bound fraction B.
· DNA
extraction
o DNA
purification with QIAquick PCR Purification Kit (Quiagen, Cat. no. 28104), following
manufacturer instructions. Step 3 and 4: Time of centrifugation : 60sec.
o Step
5: Centrifuge the column in a 2 ml collection tube for 1 min, without the cap.
o Step
7: Before the centrifugation, let the column stand for 2 minutes at room
temperature. Elution volume: 50 µl.
o In
order to obtain more DNA, it is possible to proceed to a second elution in 30
µl
o Quantify
purified DNA with the Qubit® 2.0 Fluorometer (HS DNA
assay) following manufacturer instructions.
o use
1 µl of this DNA for PCR in 25 µl reactions (quantitative real-time PCR) or 10
µl (PCR)
Ref.:
Cosseau C, Azzi A, Smith K, Freitag M, Mitta G, Grunau
C. "Native chromatin immunoprecipitation (N-ChIP) and ChIP-Seq of
Schistosoma mansoni: Critical experimental parameters." Mol Biochem
Parasitol. 2009;166:70-6. [PubMed]
Umlauf D, Goto Y, Feil R. "Site-Specific Analysis
of Histone Methylation and Acetylation" Methods Mol Biol. 2004;287:99-120.
[PubMed]
O'Neill LP, Turner BM. "Immunoprecipitation of
native chromatin: NChIP." Methods. 2003 Sep;31(1):76-82. [PubMed]